WDRR: WD40 Repeat Recognition
| [HOME] [JOB LIST] [PRE-CALCULATED RESULTS] [VISITORS] [ABOUT] |
| Pre-calculated Results - Accession: ENSDARP00000102548 [Danio rerio] [Download result as txt] [Query sequence in FASTA]
Input parameters:
Query = ENSDARP00000102548 (Len=1239) Tmplt = WD40-str (Len=40) Method = bdp Gap-open = 50 Gap-extend = 3 Database = /home/pub/blastdb/nr90 E-value = 0.001 Iteration = 3Query sequence (ENSDARP00000102548, Len=1239): (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) 10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
Query 1 MASTMIPYTVNIKLAARTLTGTLNLQNKTAVDWGWQGLIAQGCHSSILIIDPNTAQTIQVLERHKANVVKVKWSRENYHH 80
WD40-rpts (_________________WD 1_________________) (______________________WD
Query 81 SLSSPYSLRLASADAAGKIIVWDVVSGMAHCEIQEHSKPIQDMDWLWAQDASRDLLLAVHPPNYIVLWNGDTGTKLWKKS 160
WD40-rpts 2_____________________) (___________________WD 3__________________) (_______
Query 161 YAENILSFSFDPFEPSNLALLTSEGIVFITDFSHSKPPGSGGKKVYIASPHSSPAHSKPAAAQPTGAKKALNKVKVLITN 240
WD40-rpts __________WD 4________________) (_________________WD 5________________) (_
Query 241 EKPTAEAVTLNDCLQLSYLPSKRNHMLLLYPREILILDLELSQTVGVVAIERSGVPFIQVIPCAQRDALYCLHENGCITL 320
WD40-rpts ________________WD 6_________________) (_________________
Query 321 RVCRSTTPSPNETVTDPEQNSQELVYDLRSQCDAIRVTKTVRPYRVVICPVNENKAVLVVSDGRVMLWELKAHASKSSSN 400
WD40-rpts __WD 7___________________) (_________________WD 8_________________)
Query 401 LSSGLPPLYSAVNFCGTPLRQNQKCIPDLSLNSMIGHSLIPGVDSPRPLADQKEVHLKFLLTGLLSGLPLPPFSLRMCPP 480
WD40-rpts (___________________WD 9__________________) (____________
Query 481 LTTKNINHYQPLLAVGTSNGSVLVYNLTSGLLHKELSVHSCEVRGIEWISLTSFLSFATSVPNNLGLVRNELQHVDLRTG 560
WD40-rpts _____WD10________________) (_____________________WD11____________________)(___
Query 561 RCFAFRGERGNDEPAIEMIKVSHLKQYLVVVFRDKPLELWDVRTGTLLREMAKNFPTVTALEWSPSHNLKSLKKKQLAAR 640
WD40-rpts _________________WD12___________________) (_____________________WD13__________
Query 641 EAMARQTTLADAEQSSVESSVISLLQDAESKSESSQGISAREHFVFTDTDGQVYHITVEGNTVKDGARIPPDGSMGSIAC 720
WD40-rpts __________) (___________________WD
Query 721 IAWKGDTLVLGDVDGNLNFWDLKARLSRGVPTHRGWVKKIRFAPGKGNQKLLVMYTDGAEVWDTKEVQMVSSIRVGRNVN 800
WD40-rpts 14__________________) (_________________WD15_________________) (_____________
Query 801 YRILDIDWCTSDKVVLASDDGCVRVLEMAMKSASYRMDEQDLTDPVWCPYLLLPRAALTLKAFLLLQPWMDTFTMDITQV 880
WD40-rpts _____WD16_________________)
Query 881 DYKEKDEIKGLIQEQLNSLSNDIKSVLQDPNLSLLQRCLLVSRLFGDESDLQFWTVASHYIQAFAQSAQSNESVPEGQAA 960
WD40-rpts (______________________WD17__________
Query 961 ASHLDICHDILCESSFFQGFQLERVRLQEVKRSSYEHTKKCADQLLLLGQTDRAVQLLLETSADNSSYYCDSLKACLVTT 1040
WD40-rpts ____________) (_______________WD18
Query 1041 ITSSGPSQSTIKLVATNMIANGKLAEGVQLLCLIDKAADACRYLQTYGEWTRAAWLAKVRLNAAEGSDVLKRWAEHLCSP 1120
WD40-rpts ______________)
Query 1121 QVNQKSKAMLVLLSLGCFQKVGEMLHSMRYFDRAALFIEACLKYGVMETNDDINKLVGAAFVDYAKLLRSIGLKQGAVHW 1200
WD40-rpts
Query 1201 ASRAGEAGKQLLEDLSQTEGTGTESSPADDTDNSLVNIE 1239
WD40-rpts
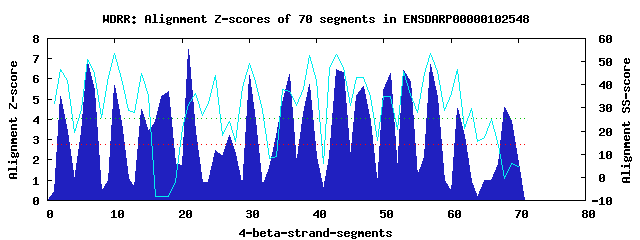 18 WD40-repeats found in QUERY=ENSDARP00000102548 (Len=1239): (Red - Z-score>4.0) Rpts Segment Boundary Z-score Score P-value Identities SS-score WD 1: Seg 2 12-51 5.14 327.17 0.00242 25.0% 46.80 WD 2: Seg 6 55-103 6.87 398.45 0.00027 32.5% 51.08 WD 3: Seg 10 107-149 5.66 348.57 0.00125 12.5% 53.73 WD 4: Seg 14 153-191 4.48 299.83 0.00566 25.0% 44.72 WD 5: Seg 18 194-232 5.40 337.79 0.00174 15.0% -8.38 WD 6: Seg 21 239-278 7.44 422.06 0.00013 15.0% 31.38 WD 7: Seg 27 303-346 3.20 246.73 0.02907 15.0% 24.28 WD 8: Seg 30 350-389 6.18 369.89 0.00064 20.0% 49.34 WD 9: Seg 34 409-451 3.23 248.05 0.02792 12.5% 8.87 WD10: Seg 36 468-506 6.20 370.63 0.00063 12.5% 37.14 WD11: Seg 39 510-556 5.72 350.95 0.00116 17.5% 52.67 WD12: Seg 43 557-601 6.46 381.67 0.00045 15.0% 52.96 WD13: Seg 47 605-651 5.62 346.74 0.00132 20.0% 43.12 WD14: Seg 53 699-741 6.41 379.65 0.00048 25.0% 45.56 WD15: Seg 57 744-783 6.72 392.29 0.00032 22.5% 53.54 WD16: Seg 61 787-827 4.53 301.74 0.00533 17.5% 46.56 WD17: Seg 68 924-973 4.57 303.32 0.00508 15.0% -0.29 WD18: Seg 69 1021-1055 3.92 276.38 0.01168 17.5% 5.82Multiple sequence alignment of 18 WD40-repeats: (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) WD 1 12-51 IK-----L---AAR-----TLTGTLNLQN----------KTAV----DWGW----Q-G--------------------LIAQGC--------HSSILIID WD 2 55-103 -A-----Q---TIQ-----VLER--HKAN----------VVKV----KWSR----E-NYHHSLSSPYSLR--------LASADA--------AGKIIVWD WD 3 107-149 -G-----M---AHC-----EIQE--HSKP----------IQDM----DWLWAQDAS-R----------DL--------LLAVHP--------PNYIVLWN WD 4 153-191 -G-----T---KLW-----KKSY--AEN-----------ILSF----SFDP---FE-P----------SN--------LALLTS--------EGIVFITD WD 5 194-232 -H-----S---KPP-----GSGG--KKVY----------IASP----HSSP----A-H----------SK--------PAAAQP--------TGAKKALN WD 6 239-278 TN-----E---KPT-----AEAV--TLND----------CLQL----SYLP----S-K----------RN--------HMLLLY--------PREILILD WD 7 303-346 CA-----Q---RDA-----LYCL--HENG----------CITLRVCRSTTP----S-P----------NE--------TVTDPE--------QNSQELVY WD 8 350-389 -S-----Q---CDA-----IRVT--KTVR----------PYRV----VICP----V-N---------ENK--------AVLVVS--------DGRVMLWE WD 9 409-451 YS-----AVNFCGT-----PLRQ--NQKC----------IPDL----SLNS----M-I----------GH--------SLIPGV--------DSPRPLAD WD10 468-506 -L-----P---LPP-----FSLR--MCPP----------LTTK----NINH----Y-Q----------PL--------LAVGTS--------NGSVLVYN WD11 510-556 -G-----L---LHK-----ELSV--HSCE----------VRGI----EWIS----L-T----------SF--------LSFATSVPNNLGLVRNELQHVD WD12 557-601 LRTGRCFA---FRG-----ERGN--DEPA----------IEMI----KVSH----L-K----------QY--------LVVVFR--------DKPLELWD WD13 605-651 -G-----T---LLR-----EMAK--NFPT----------VTAL----EWSP----S-H----------NLKSLKKKQLAAREAM--------ARQTTLAD WD14 699-741 EG-----N---TVKDGARIPPDG--SMGS----------IACI----AWKG----D-T--------------------LVLGDV--------DGNLNFWD WD15 744-783 -A-----R---LSR-----GVPT--HRGW----------VKKI----RFAP----GKG----------NQ--------KLLVMY--------TDGAEVWD WD16 787-827 -V-----Q---MVS-----SIRV--GRN-----------VNYR----ILDI----D-W-------CTSDK--------VVLASD--------DGCVRVLE WD17 924-973 LF-----G---DES-----DLQF--WTVASHYIQAFAQSAQSN----ESVP----E-G----------QA--------AASHLD--------ICHDILCE WD18 1021-1055 T------------------SADN--SSYY----------CDSL----KACL----V-T----------TI--------TSSGPS--------QSTIKLVA WD40 Template TG-----E---CLR-----TLSG--HTSG----------VTSV----SFSP----D-G----------KR--------LVSGSE--------DGTIKVWD |
|
Copyright © 2012-2015 Ziding Zhang's Lab, China Agricultural University Powered and maintained by Chuan Wang Last modified: 09 Jan 2019 |