WDRR: WD40 Repeat Recognition
| [HOME] [JOB LIST] [PRE-CALCULATED RESULTS] [VISITORS] [ABOUT] |
| Pre-calculated Results - Accession: ENSDARP00000101591 [Danio rerio] [Download result as txt] [Query sequence in FASTA]
Input parameters:
Query = ENSDARP00000101591 (Len=937) Tmplt = WD40-str (Len=40) Method = bdp Gap-open = 50 Gap-extend = 3 Database = /home/pub/blastdb/nr90 E-value = 0.001 Iteration = 3Query sequence (ENSDARP00000101591, Len=937): (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) 10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
Query 1 MKFAYKFSNLLGAFYRQGNISFSKNGDSVISPVGNRISVFDLKNNKSETLPVSTSKNIACFDISPDGNLAILIDEDGAAI 80
WD40-rpts (_________________WD 1________________) (_________________WD 2______________
Query 81 LVSLVTRAVLHHHHFHNPVHNVSFSPDGRKFVVTKENVALMYHAPGRNKEFNAFALNKTFYGPFDEMTCIDWTDDSKCFA 160
WD40-rpts __) (________________WD 3________________)(_____________________WD 4__________
Query 161 VGSKDMTTWIFGAERWANLIYYSLGGHKDIIVGCFFEKDSLDMYTVSQEGTLCIWESDTDLEGLRKGPKYAERKEKEQKK 240
WD40-rpts ___________) (_________________WD 5________________) (___
Query 241 LRVLEMEEDEANDIEIMGEDGETRGDVIKGSAETPEEVEGIKNVRYLQKSKYYFNKEGDFNKLTAAAFHKRTHILVTGFA 320
WD40-rpts ________________WD 6___________________) (_________________WD 7_________
Query 321 SGAFHLHELPGFDLIHSLSISDQRISAISLNPTGDWISFGCSGLGQLLVWEWQSESYVFKQQGHFNNMNSLAYSPDGQYL 400
WD40-rpts _______) (_________________WD 8_________________) (_________________WD 9____
Query 401 ATGGDDGKVKVWNTNSGLCFVTFTEHSSGVTEVAFTSSGFVVVSASLDGTVRAFDLHRYRNFRTMTSPRPAQFSSLAVDG 480
WD40-rpts ____________) (_________________WD10________________)(___________________WD11_
Query 481 SGELVCAGSQDSFEIFLWSMQTGRLLEVLSGHEGPVSNLCFSPVQSVLASVSWDKTVRLWDMLDSWQTKETLQLTSDGLA 560
WD40-rpts __________________) (_________________WD12________________) (_______________
Query 561 VTYHPNGMELAVASLDGEITFWNPQTGRQTGSITGRHDLQMGRKESEKITAKQSAKGKSFTTLCYSADGESILTGGKSKF 640
WD40-rpts __WD13________________) (______________WD14_________
Query 641 VCIYNIKEHLLMKKFEISCNLSLDAMEEFLDRRKMTEFGSLSLVDEGTGDGNGVELSLPGVKKGDMSTRCFKPEISVSSL 720
WD40-rpts ____) (________________
Query 721 RFSPTGRSWAAASTEGLLIYSLDAALVFDPYDLDIDVTPTSIRQQITKKEWASAILLSFRLNEILLIREVLEAIPFDQIT 800
WD40-rpts WD15________________)
Query 801 VVCSSLPDVYVDKLLNFVASTLEKSNHLQFYLTWAQCLLTLHGQKLKNRPGFVLPTVQQLLKSIQRHSDDLSKLCDWNIY 880
WD40-rpts
Query 881 NIRYAAALAKQRGLKRTASEIYGAEKEDSSEAMDSEGDEENMTNEPIMLENAIQLSD 937
WD40-rpts
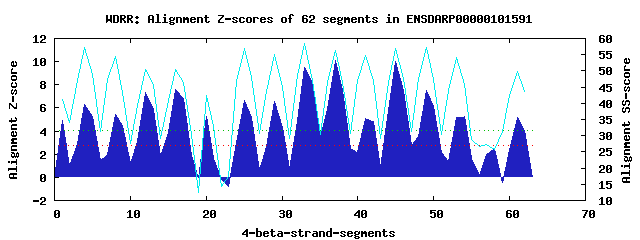 15 WD40-repeats found in QUERY=ENSDARP00000101591 (Len=937): (Red - Z-score>4.0) Rpts Segment Boundary Z-score Score P-value Identities SS-score WD 1: Seg 1 3-41 4.90 317.24 0.00330 25.0% 41.20 WD 2: Seg 4 45-83 6.27 373.48 0.00058 17.5% 57.26 WD 3: Seg 8 87-124 5.45 339.60 0.00165 30.0% 54.45 WD 4: Seg 12 125-172 7.30 416.11 0.00015 25.0% 50.33 WD 5: Seg 16 178-216 7.63 429.85 0.00010 22.5% 50.54 WD 6: Seg 20 237-280 5.30 333.55 0.00199 20.0% 42.35 WD 7: Seg 25 290-328 6.67 390.38 0.00034 17.5% 56.86 WD 8: Seg 29 332-371 6.56 385.81 0.00039 25.0% 55.08 WD 9: Seg 33 375-413 9.48 506.20 9.4e-06 35.0% 58.59 WD10: Seg 37 417-455 10.12 532.57 4.1e-06 45.0% 56.40 WD11: Seg 41 456-499 5.11 325.70 0.00254 20.0% 54.82 WD12: Seg 45 503-541 10.02 528.70 4.7e-06 42.5% 56.88 WD13: Seg 49 545-583 7.51 424.92 0.00012 27.5% 57.38 WD14: Seg 54 613-645 5.25 331.49 0.00212 15.0% 45.87 WD15: Seg 61 704-741 5.15 327.22 0.00242 22.5% 49.70Multiple sequence alignment of 15 WD40-repeats: (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) WD 1 3-41 FAY--------KFSNLLGAFYRQGNIS-FSKNGDS---VISP-VGN--RISVFD WD 2 45-83 -NK--------SETLPVSTSKNIACFD-ISPDGNL---AILI-DEDG-AAILVS WD 3 87-124 -RA--------VLHHHHFHNP-VHNVS-FSPDGRK---FVVT-KENV-ALMYHA WD 4 125-172 PGRNKEFNAFALNKTFYGPFDEMTCID-WTDDSKC---FAVG-SKDM-TTWIFG WD 5 178-216 -NL--------IYYSLGGHKDIIVGCF-FEKDSLD---MYTV-SQEG-TLCIWE WD 6 237-280 EQK--------KLRVLEMEEDEANDIEIMGEDGETRGDVIKG-SAET-PEEVEG WD 7 290-328 -SK--------YYFNKEGDFNKLTAAA-FHKRTHI---LVTG-FASG-AFHLHE WD 8 332-371 -FD--------LIHSLSISDQRISAIS-LNPTGDW---ISFGCSGLG-QLLVWE WD 9 375-413 -ES--------YVFKQQGHFNNMNSLA-YSPDGQY---LATG-GDDG-KVKVWN WD10 417-455 -GL--------CFVTFTEHSSGVTEVA-FTSSGFV---VVSA-SLDG-TVRAFD WD11 456-499 LHR-----YRNFRTMTSPRPAQFSSLA-VDGSGEL---VCAG-SQDSFEIFLWS WD12 503-541 -GR--------LLEVLSGHEGPVSNLC-FSPVQSV---LASV-SWDK-TVRLWD WD13 545-583 -SW--------QTKETLQLTSDGLAVT-YHPNGME---LAVA-SLDG-EITFWN WD14 613-645 ---------------QSAKGKSFTTLC-YSADGES---ILTG-GKSK-FVCIYN WD15 704-741 -GD--------MSTRCFKPEISVSSLR-FSPTGRS---WAAA-STEG-LL-IYS WD40 Template TGE--------CLRTLSGHTSGVTSVS-FSPDGKR---LVSG-SEDG-TIKVWD |
|
Copyright © 2012-2015 Ziding Zhang's Lab, China Agricultural University Powered and maintained by Chuan Wang Last modified: 09 Jan 2019 |