WDRR: WD40 Repeat Recognition
| [HOME] [JOB LIST] [PRE-CALCULATED RESULTS] [VISITORS] [ABOUT] |
| Pre-calculated Results - Accession: ENSDARP00000101029 [Danio rerio] [Download result as txt] [Query sequence in FASTA]
Input parameters:
Query = ENSDARP00000101029 (Len=1843) Tmplt = WD40-str (Len=40) Method = bdp Gap-open = 50 Gap-extend = 3 Database = /home/pub/blastdb/nr90 E-value = 0.001 Iteration = 3Query sequence (ENSDARP00000101029, Len=1843): (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) 10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
Query 1 MSSKALWQQPYVNIFKHMKIEEWKKVSKEGDVSLYMDKNLKCSVFSIRGSIQASNYILLPKTSSQTLGLTGRYLYLLFKP 80
WD40-rpts
Query 81 SPNKHFIVHLDIAAEEGQVIRVSFSNLFKEFKSTATWLQFPFLCGAAHGSVDENTSTTAKQGLVGPAPWSTRWTCLTMDL 160
WD40-rpts
Query 161 RYVLSVYLNRTHSHLKSIKLCANMSVKNIITSDLLFDPGLSFSEFRQSGVMLPDGSAPMPREMCFPVPKGQSWHHLYDYI 240
WD40-rpts
Query 241 RFPSDGIKSPIDFIQKASNSAAKKASAEKESPQREESRMVRISKPVNDRVSLVKQITKPKRRSTKQCSEIQTPSLPKLIY 320
WD40-rpts
Query 321 SPSRVSESPGSPGQRSQAGEEVTVVFADDAGVHVYASCEDDIYIHTTDSEEEITVIDAASPRPVSLSSDRSVKQSQKLYP 400
WD40-rpts
Query 401 DPILKLNRIIGFGGATMRCAVWSLDGEEVVYPCHAIIVSMTVSSGQQRFFIGHTDKVSALALNASSTVLASAQTGTNSLI 480
WD40-rpts (________________WD 1________________)(__________________WD 2_______________
Query 481 RLWSYSAGVCQTIIPTHTHALNLLSFSHSGGVLCGVGKDSHNKTTVVLWNTRHVTDGSVVPVLAKAHTDVDIQTIKIAFF 560
WD40-rpts ___) (___________________WD 3__________________)
Query 561 DDTRMVSCGSGNVRLWRVRGGSLRSCPVNLGQYHNLEFTDLAFEQGHTQNRPVDDRTLFVSSRSGHILEIDYVAVAIKNI 640
WD40-rpts (_______________WD 4______________)
Query 641 RRLLPAHESHTHQREKQTFSTGAGIAVNSISVCAKFCVTGSEDGFMRLWPLDFSSVFLEAEHEGPVSLVNVSSDGSRVLA 720
WD40-rpts (_________________WD 5________________) (_________________WD 6______
Query 721 ATSTGNLGYLDVASRGYRTLMRSHTASVLGFSVDGVRRRVTTVSRDATVRVWDMDTLQQLYDFVTDDDENPCSVAFHPSM 800
WD40-rpts __________) (_________________WD 7________________)(___________________WD 8___
Query 801 PLFACGSSSGAVRVFDIQTSSLIAQHRQHCGSVECVIYSPDGQCLFSAGSGYLVLYSSSEREHHVVRVLGNVVMCGVNRS 880
WD40-rpts _______________) (_________________WD 9________________) (__________
Query 881 PDILAVSSDSRRLALIGPTEYIVTIMEARSLNELLRVDVSIVDVESSTLDSALNACFSPASLSHLLISTSANKILWIDTH 960
WD40-rpts ______WD10________________) (_________________WD11________________)
Query 961 TGQLLRQVSEVHKHQCESLAVSEDGHYLLTAGQNSLKVWDYDMKLDVNSQVFIGHSEPIRQVRFTPDQVGVVTAGDALFF 1040
WD40-rpts (_________________WD12________________)(__________________WD13_________________
Query 1041 WDFLAPPQQSEHCRSPALNVSSRKSAQSWAGLNISELSNEKPREALFHPSSPPVMNFLSTKRNDTEDLLCKSAAELQLSP 1120
WD40-rpts _) (_________________WD14________________) (_________________W
Query 1121 SADHAHSPPPRHASFLRVTEWEKDEDTPTLSGHQSGSEKTQGHTLIHPDCYKHFTPRFKTSVVDQSVVASPSGEAGLSLK 1200
WD40-rpts D15________________) (____
Query 1201 AVIGYNGNGRSNMVWNPDTGLFVYTCGSIVIIEDLHTGSQKQWFGHTEEISTLAVTHDALTVASASGGGASHSSLICIWN 1280
WD40-rpts _____________WD16________________)(____________________WD17____________________)
Query 1281 VQDGSCRHKLAYHNGAVQSLTFSRDDMYLLSVGDFSEALVALWSTRSFELLCTVQLSIPLHDAAFSPFTANELMLVGSSA 1360
WD40-rpts (__________________WD18_________________)(___________________WD19____________
Query 1361 VVFTRIHTHDNTTELQVQKVGLPDAVGQAEVTSVCYNTRNILYTGTNSGHVCVWDCNTQRCFVTWEADRGEIGVLVCCGN 1440
WD40-rpts _______) (_______________WD20______________) (________________WD21_
Query 1441 MLVTGSNTKKLRLWSVASVQSLSNSDSKRSRAQDDGTQVQLEQELLLDGTIVSAAFDKVMDMGIVGTTAGTLWYINWTDS 1520
WD40-rpts ______________) (________________________WD22_______________________) (
Query 1521 TSIRLISGHKSKVNGVVCSPDEQHFVTCGQDGSVRVWTLQSHELLVQFQVLNQSCECVCWSPPRGVCGSRGRMCVAAGYS 1600
WD40-rpts _________________WD23________________) (_____________________WD24_____________
Query 1601 DGTVRIFSLDSAEMELKLQPQPDSVCALQFSHYGDVLLSGARDGVITVCSPVTGVSIRTITDHRGASINTLQCTSKQFKE 1680
WD40-rpts _______) (_________________WD25________________) (___________________W
Query 1681 FGLDGNELWLAVSADRRVSIWAADWTDNKCELLDWLTFPAPSPREDLHTLPPSLAAFSPSQRGVVVYTGYAMEKEIIFYS 1760
WD40-rpts D26__________________) (__________________WD27__________________)
Query 1761 LYKKQIVRTISLSDWALSLCLSGKLLAVGSNQRVLNLIHSSSGRFQDFTSHSDSVQVCSFSISGSQLFTAAHNQLLLWTV 1840
WD40-rpts (_________________WD28________________)(_________________WD29_________________)
Query 1841 NGL 1843
WD40-rpts
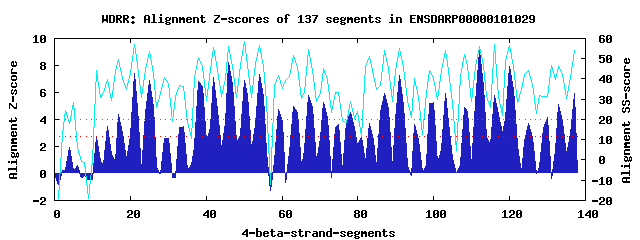 29 WD40-repeats found in QUERY=ENSDARP00000101029 (Len=1843): (Red - Z-score>4.0) Rpts Segment Boundary Z-score Score P-value Identities SS-score WD 1: Seg 17 405-442 4.39 295.89 0.00639 12.5% 49.57 WD 2: Seg 21 443-484 7.44 422.15 0.00013 22.5% 57.22 WD 3: Seg 25 488-530 6.90 399.80 0.00025 30.0% 53.34 WD 4: Seg 34 588-622 3.48 258.22 0.02044 17.5% 36.14 WD 5: Seg 38 652-690 6.83 396.94 0.00028 27.5% 50.56 WD 6: Seg 42 693-731 7.12 408.70 0.00019 22.5% 55.38 WD 7: Seg 46 735-773 8.25 455.63 4.5e-05 27.5% 56.17 WD 8: Seg 50 774-816 6.90 399.83 0.00025 22.5% 58.40 WD 9: Seg 54 820-858 7.35 418.17 0.00014 22.5% 56.71 WD10: Seg 59 870-907 4.72 309.43 0.00420 15.0% 41.54 WD11: Seg 63 919-957 4.96 319.68 0.00306 15.0% 51.45 WD12: Seg 67 962-1000 5.79 353.82 0.00106 35.0% 54.78 WD13: Seg 71 1001-1042 5.27 332.26 0.00207 25.0% 45.24 WD14: Seg 75 1061-1099 3.60 263.50 0.01737 10.0% 33.38 WD15: Seg 78 1102-1140 4.50 300.72 0.00550 12.5% 28.71 WD16: Seg 83 1196-1234 3.70 267.44 0.01539 20.0% 51.02 WD17: Seg 87 1235-1280 5.96 360.80 0.00085 20.0% 53.99 WD18: Seg 91 1284-1324 7.27 415.14 0.00016 30.0% 55.28 WD19: Seg 95 1325-1368 3.66 265.92 0.01613 20.0% 40.35 WD20: Seg100 1381-1415 5.25 331.37 0.00213 25.0% 40.22 WD21: Seg103 1419-1455 5.90 358.44 0.00092 20.0% 54.23 WD22: Seg108 1464-1516 4.89 316.84 0.00334 15.0% 52.26 WD23: Seg112 1520-1558 9.12 491.45 1.5e-05 37.5% 56.29 WD24: Seg116 1562-1608 5.80 354.35 0.00104 22.5% 57.16 WD25: Seg120 1612-1650 7.92 441.78 6.9e-05 32.5% 56.69 WD26: Seg125 1660-1702 3.68 266.66 0.01576 20.0% 44.34 WD27: Seg130 1719-1760 4.13 285.17 0.00890 17.5% 31.35 WD28: Seg133 1761-1799 5.13 326.45 0.00248 15.0% 46.09 WD29: Seg137 1800-1839 5.90 358.34 0.00092 20.0% 54.22Multiple sequence alignment of 29 WD40-repeats: (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) WD 1 405-442 -----K-------------L-N----RIIG-FGG-ATMRCA---------VWSL---D--------GEEV-V---YPC-----H--AI----IVSMTV WD 2 443-484 SS---G-------------Q-Q----RFFI-GHT-DKVSAL---------ALNA---S--------STVL-A---SAQ---TGT--NS----LIRLWS WD 3 488-530 -G---V-------------C-Q----TIIP-THT-HALNLL---------SFSH---S--------GGVL-C---GVG-----K--DSHNKTTVVLWN WD 4 588-622 ---------------------V----NLGQ-YHN-LEFTDL---------AFE----Q--------GHTQ-N---RPV-----D--DR----TLFVSS WD 5 652-690 -H---Q-------------R-E----KQTF-STG-AGIAVN---------SISV---C--------AKFC-V---TGS-----E--DG----FMRLWP WD 6 693-731 -F---S-------------S-V----FLEA-EHE-GPVSLV---------NVSS---D--------GSRV-L---AAT-----S--TG----NLGYLD WD 7 735-773 -R---G-------------Y-R----TLMR-SHT-ASVLGF---------SVDG---V--------RRRV-T---TVS-----R--DA----TVRVWD WD 8 774-816 MDTLQQ-------------L-Y----DFVT-DDD-ENPCSV---------AFHP---S--------MPLF-A---CGS-----S--SG----AVRVFD WD 9 820-858 -S---S-------------L-I----AQHR-QHC-GSVECV---------IYSP---D--------GQCL-F---SAG-----S--GY----LVLYSS WD10 870-907 GN---V---------------------VMC-GVN-RSPDIL---------AVSS---D--------SRRL-A--LIGP-----T--EY----IVTIME WD11 919-957 -V---S-------------I-V----DVES-STL-DSALNA---------CFSP---A--------SLSH-L---LIS-----T--SA----NKILWI WD12 962-1000 -G---Q-------------L-L----RQVSEVHK-HQCESL---------AVSE---D--------GHYL-L---TAG--------QN----SLKVWD WD13 1001-1042 YD---M-------------K-LDVNSQVFI-GHS-EPIRQV---------RFTP---D--------QVGV-V---TAG-----D--------ALFFWD WD14 1061-1099 -S---S-------------R-K----SAQS-WAG-LNISEL---------SNEK---P--------REAL-F---HPS-----S--PP----VMNFLS WD15 1102-1140 -R---N-------------D-T----EDLL-CKS-AAELQL---------SPSA---D--------HAHS-P---PPR-----H--AS----FLRVTE WD16 1196-1234 -G---L-------------S-L----KAVI-GYNGNGRSNM---------VWNP---D--------TGLF-V---YTC--------GS----IVIIED WD17 1235-1280 LH---T------------GS-Q----KQWF-GHT-EEISTL---------AVTH---D--------ALTV-A---SASGGGASH--SS----LICIWN WD18 1284-1324 -G---S-------------C-R----HKLA-YHN-GAVQSL---------TFSR---D--------DMYL-L---SVG-----DFSEA----LVALWS WD19 1325-1368 TR---S-------------FEL----LCTV-QLS-IPLHDA---------AFSPFTAN--------ELML-V---GSS-----A--VV----FTRIHT WD20 1381-1415 ---------------------G----LPDA-VGQ-AEVTSV---------CYNT---R--------N-IL-Y---TGT-----N--SG----HVCVWD WD21 1419-1455 -Q---R-------------C-F----VTWE-ADR-GEIGVL---------VCC-------------GNML-V---TGS-----N--TK----KLRLWS WD22 1464-1516 NS---DSKRSRAQDDGTQVQ-L----EQEL-LLD-GTIVSA---------AFDK---V--------MDMG-I---VGT-----T--AG----TLWYIN WD23 1520-1558 -S---T-------------S-I----RLIS-GHK-SKVNGV---------VCSP---D--------EQHF-V---TCG-----Q--DG----SVRVWT WD24 1562-1608 -H---E-------------L-L----VQFQ-VLN-QSCECV---------CWSP---PRGVCGSRGRMCV-A---AGY-----S--DG----TVRIFS WD25 1612-1650 -A---E-------------M-E----LKLQ-PQP-DSVCAL---------QFSH---Y--------GDVL-L---SGA-----R--DG----VITVCS WD26 1660-1702 ----------------------------IT-DHR-GASINTLQCTSKQFKEFGL---D--------GNELWL---AVS-----A--DR----RVSIWA WD27 1719-1760 -P---A-------------P-S----PRED-LHT-LPPSLA---------AFSP---S--------QRGV-VVYTGYA-----M--EK----EIIFYS WD28 1761-1799 LY---K---------------K----QIVR-TIS-LSDWAL---------SLCL---S--------GKLL-A---VGS-----N--QR----VLNLIH WD29 1800-1839 SS---S------------GR-F----QDFT-SHS-DSVQVC---------SFSI---S--------GSQL-F---TAA-----H--N-----QLLLWT WD40 Template TG---E-------------C-L----RTLS-GHT-SGVTSV---------SFSP---D--------GKRL-V---SGS-----E--DG----TIKVWD |
|
Copyright © 2012-2015 Ziding Zhang's Lab, China Agricultural University Powered and maintained by Chuan Wang Last modified: 09 Jan 2019 |