WDRR: WD40 Repeat Recognition
| [HOME] [JOB LIST] [PRE-CALCULATED RESULTS] [VISITORS] [ABOUT] |
| Pre-calculated Results - Accession: ENSDARP00000100360 [Danio rerio] [Download result as txt] [Query sequence in FASTA]
Input parameters:
Query = ENSDARP00000100360 (Len=1908) Tmplt = WD40-str (Len=40) Method = bdp Gap-open = 50 Gap-extend = 3 Database = /home/pub/blastdb/nr90 E-value = 0.001 Iteration = 3Query sequence (ENSDARP00000100360, Len=1908): (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) 10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
Query 1 SLALHPERVLVATGQVGKEPYICVWDSYTVQTVSILRDMHTHGIACLAFDLDGQCLVSVGLDSKNTICVWDWRQGKVLAA 80
WD40-rpts (__________________WD 1_________________) (_____
Query 81 AQGCIGKIFDISWDLYQQSKLVSCGVKHIKFWSLCGNALTPKRGVFGKTGDLQTILCLACARDEITYSGALNGDIYVWKG 160
WD40-rpts ____________WD 2________________) (________________WD 3_______________)(
Query 161 INLIRTVQGAHGVKIFACHKGFATGGRDGCIRLWDLNFKPITVIDLRETDQGYKGLSVRSVCWRGDHILVGTQDSEIFEV 240
WD40-rpts _______________WD 4_______________)
Query 241 VVHDRTKPFLIMQGHCEGELWALAVHPTKPLAMTGSDDRSVRIWSLVDHALIARCNMEEPIRCAAVSTDGIHLALGMKDG 320
WD40-rpts (_________________WD 5_________________) (_________________WD 6___________
Query 321 SFTVLRVRDMTEVVHIKDRKEAIHELKYSPDGAHLAVGSNDNSVDIYGVVQRYKKVGECVGSSSFITHMDWSTDSKYLQT 400
WD40-rpts _____) (_________________WD 7________________) (_________________WD 8______
Query 401 NDGNGRRLFYRMPSGKEVTNREELKLVQWSSWTCVLGPEVNGIWPKYSDINDINSVDANFSSQVLVTADDYGLVKLFRYP 480
WD40-rpts __________) (_________________WD 9________________)
Query 481 CIRKGAKFKKYLGHSAHITNIRWSHDYQWVITIGGADHSVFQWKFVPERKSKDTLHIAPQETLIDSHSEESDSDQSDVPE 560
WD40-rpts (_________________WD10_________________) (_________________WD11________
Query 561 MDSEIEQETQLTYRRQVYKEDLPQLKEQCKEKHRAMAMKKRERAPASGMRLHFIHGYRGYDCRSNLFYTQTGEIVYHVAA 640
WD40-rpts ________)
Query 641 VGVVYNRQQNTQRFYLGHDDDILCLAIHPVKDYVATGQVGRDSSIHIWETEFLKPLSVLKGFHQFGVCALDFSAADGKRL 720
WD40-rpts (__________________WD12_________________) (___________________WD13____
Query 721 ASVGLDDNHTIVLWEWRKGEKLSTIRGSKDKIFVIKINPYMPDKLISAGVKHMKFWHRAGGGLIGKKGSMGKTETMMCAV 800
WD40-rpts ______________) (_________________WD14________________)(___________________WD1
Query 801 YGWTEEMVFSGTCTGDICIWRDMFLVKTVKAHDGPVFSMHALEKGFVTGGKDGIVALWDDTFERCLKTYAIKRAVLAPGS 880
WD40-rpts 5___________________)(________________WD16________________) (___
Query 881 KGLLLEDNPSIRAISLGHGHILVGTKNGEILEVDKNGPITLLVQGHMEGEVWGLATHPHLPLCATVSDDKTLRIWDLSPS 960
WD40-rpts _____________WD17________________) (_________________WD18________________) (
Query 961 HCMLAVRKLKKGGRCCCFSPDGKALAVGLNDGSFLIVNADTLEDLVSFHHRKDIISEIRFSPVTGKYLAVASGDSFVDIY 1040
WD40-rpts _________________WD19________________) (_________________WD20_________________
Query 1041 NVMSSKRVGVCKGSMNYITHLDWDKRVGKLLQVNTSAKEQLFFEAPRGKKQSIPVTEVEKIDWSTWTCVLGSSCEGIWPV 1120
WD40-rpts ) (_________________WD21_________________) (______
Query 1121 VSEVTEVTAACLSYDKKLLATGDDLGYVKLFKYPVKGKYAKFKRYMAHSTHVTNVRWSHDDSLLASVGGSDTCLMIWNHE 1200
WD40-rpts ___________WD22________________) (_________________WD23_________________)
Query 1201 TESCREARQCDSEESDIESEDDGGYDSDVTRENEIVYVIKALSTNMRPMTGVKPHLQQKEPTVDESDGFLEMCRPPVSRA 1280
WD40-rpts (_________________WD24________________) (____________________WD25_______
Query 1281 PPMPEKLQTNNVGKKKRPIEDLVLELVFGYRGNDCRNNVHYLNEGADIIYHTASIGVVLNLTTACQSFYLEHSDDILCLT 1360
WD40-rpts _____________)(_____________________WD26____________________) (________
Query 1361 INQHPKFPNVVATGQVGDTGDMSATSPSIHVWEAMNKQTLSVLRCHHSRGVCSVSFSATGKLLLSVGLDPQHTLTIWKWQ 1440
WD40-rpts __________WD27__________________) (__________________WD28__________________)
Query 1441 EGAKVASRAGHTQRIFVAEFRPDSDTQFVSVGIKHVRFWTLAGRALLSKKGVLSSIEDARMQTMLSVAFGANNLTFTGTI 1520
WD40-rpts (_________________WD29________________) (________________WD30_________
Query 1521 SGDVCVWKEHILVRIVAKAHTGPVFTMYTTLRDGLIVTGGKERPSKEGGALKLWDQELKRCRAFRLETGQVIDCVRSVCR 1600
WD40-rpts _______)(_____________________WD31____________________)(___________________WD32_
Query 1601 GKGKILVGTRNAEIIEVGEKNAACNILVNGHMDGPVWGLGAHPTRDVFLSAAEDGTVRLWDISQRKMLNKVNLGHPARTV 1680
WD40-rpts __________________) (_________________WD33_________________)(__________________
Query 1681 SYSPEGDMVAIGMKNGEFIILLVTSLKIWGKKRDRRSAIQDIRFSPDSRYLAVGSSENAVDFYDLTLGPQLNRINCCRDI 1760
WD40-rpts WD34_________________) (_________________WD35________________) (______
Query 1761 PSFVMQMDFSADSCFVQLSTGAYKRVVYEVPSGKQVTEQAVIDRITWATWTSVLGDEVVGIWSRNTDKADVTCACVSHSG 1840
WD40-rpts _________WD36_______________) (__________________WD37___
Query 1841 INIVTGDDFGMVKLFDFPCPEKFAKHKRFLGHSAHLTNVRFTNGDRFVVSAGGDDRSLFVWRCVHTPH 1908
WD40-rpts _______________) (_________________WD38_________________)
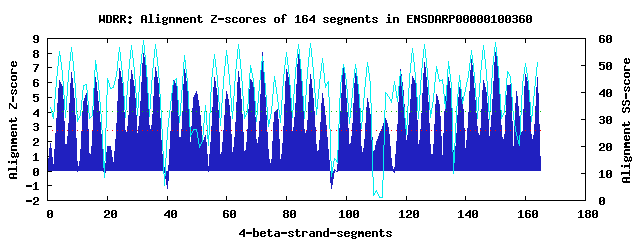 38 WD40-repeats found in QUERY=ENSDARP00000100360 (Len=1908): (Red - Z-score>4.0) Rpts Segment Boundary Z-score Score P-value Identities SS-score WD 1: Seg 4 31-71 6.16 369.28 0.00066 37.5% 55.21 WD 2: Seg 8 75-113 6.67 390.33 0.00034 17.5% 56.83 WD 3: Seg 13 123-159 5.28 332.91 0.00203 22.5% 43.13 WD 4: Seg 16 160-195 6.34 376.47 0.00053 27.5% 51.96 WD 5: Seg 24 246-285 6.98 402.96 0.00023 20.0% 56.70 WD 6: Seg 28 288-326 6.83 396.85 0.00028 22.5% 57.41 WD 7: Seg 32 330-368 7.99 444.52 6.3e-05 20.0% 59.17 WD 8: Seg 36 373-411 7.06 406.47 0.00021 20.0% 57.91 WD 9: Seg 42 440-478 6.14 368.47 0.00067 15.0% 44.13 WD10: Seg 46 485-524 6.90 399.87 0.00025 22.5% 53.67 WD11: Seg 50 531-569 5.40 337.91 0.00174 12.5% 26.39 WD12: Seg 56 649-689 6.36 377.43 0.00051 20.0% 53.96 WD13: Seg 60 693-735 5.38 336.78 0.00180 45.0% 55.39 WD14: Seg 64 739-777 6.76 394.10 0.00030 20.0% 57.67 WD15: Seg 68 778-821 6.24 372.35 0.00060 17.5% 50.15 WD16: Seg 72 822-859 8.03 446.54 6.0e-05 27.5% 53.29 WD17: Seg 77 877-914 4.20 288.10 0.00813 15.0% 38.23 WD18: Seg 80 918-956 6.89 399.17 0.00026 20.0% 54.78 WD19: Seg 84 960-998 7.91 441.59 7.0e-05 30.0% 57.88 WD20: Seg 88 1002-1041 6.55 385.27 0.00040 25.0% 58.25 WD21: Seg 92 1045-1084 5.29 333.03 0.00202 12.5% 52.65 WD22: Seg 99 1114-1152 7.17 410.89 0.00018 25.0% 50.55 WD23: Seg103 1159-1198 6.64 388.81 0.00036 27.5% 50.40 WD24: Seg107 1201-1239 4.93 318.34 0.00319 15.0% 51.06 WD25: Seg112 1249-1294 2.96 236.90 0.03924 10.0% 1.05 WD26: Seg113 1295-1341 3.57 262.08 0.01815 7.5% 40.12 WD27: Seg118 1352-1393 6.90 399.77 0.00025 17.5% 47.92 WD28: Seg122 1397-1438 6.40 379.09 0.00048 42.5% 56.36 WD29: Seg126 1442-1480 7.36 418.48 0.00014 20.0% 57.77 WD30: Seg130 1491-1528 5.18 328.59 0.00232 20.0% 49.52 WD31: Seg134 1529-1575 6.34 376.58 0.00052 25.0% 51.59 WD32: Seg138 1576-1619 5.68 349.09 0.00123 20.0% 46.13 WD33: Seg142 1622-1661 7.55 426.49 0.00011 32.5% 55.56 WD34: Seg146 1662-1702 6.17 369.56 0.00065 17.5% 57.40 WD35: Seg150 1706-1744 8.03 446.49 6.0e-05 22.5% 58.70 WD36: Seg155 1754-1789 5.88 357.40 0.00095 15.0% 46.02 WD37: Seg160 1815-1856 6.53 384.27 0.00041 25.0% 50.79 WD38: Seg164 1863-1902 6.33 376.21 0.00053 27.5% 51.09Multiple sequence alignment of 38 WD40-repeats: (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) WD 1 31-71 -Q------T----VSI--LRD-M-------HTHG-IACLAFDL-D-GQCLVS-V--GLD----SKN--TICVWD WD 2 75-113 -G------K----VLA--AAQ-G-------CIGK-IFDISWDL-Y-QQSKLV-S--CGV------K--HIKFWS WD 3 123-159 RG------------VF--GKT-G-------DLQT-ILCLACAR-D-E-ITYS-G--ALN------G--DIYVWK WD 4 160-195 GI------N----LIR--TVQ-G-------A-----HGVKIFA-C-HKGFAT-G--GRD------G--CIRLWD WD 5 246-285 -T------K----PFL--IMQ-G-------HCEGELWALAVHP-T-KPLAMT-G--SDD------R--SVRIWS WD 6 288-326 -D------H----ALI--ARC-N-------MEEP-IRCAAVST-D-GIHLAL-G--MKD------G--SFTVLR WD 7 330-368 -M------T----EVV--HIK-D-------RKEA-IHELKYSP-D-GAHLAV-G--SND------N--SVDIYG WD 8 373-411 -Y------K----KVG--ECV-G-------SSSF-ITHMDWST-D-SKYLQT-N--DGN------G--RRLFYR WD 9 440-478 -V------N----GIW--PKY-S-------DIND-INSVDANF-S-SQVLVT-A--DDY------G--LVKLFR WD10 485-524 -G------A----KFK--KYL-G-------HSAH-ITNIRWSH-D-YQWVITIG--GAD------H--SVFQWK WD11 531-569 -S------K----DTL--HIA-P-------QETL-IDSHSEES-D-SDQSDV-P--EMD------S--EIEQET WD12 649-689 -Q------N----TQR--FYL-G-------HDDD-ILCLAIHP-V-KDYVAT-GQVGRD------S--SIHIWE WD13 693-735 -L------K----PLS--VLK-G------FHQFG-VCALDFSAAD-GKRLAS-V--GLD------DNHTIVLWE WD14 739-777 -G------E----KLS--TIR-G-------SKDK-IFVIKINP-Y-MPDKLI-S--AGV------K--HMKFWH WD15 778-821 RA------GGGLIGKK--GSM-G-------KTET-MMCAVYGW-T-EEMVFS-G--TCT------G--DICIWR WD16 822-859 DM------F----LVK--TVK-A-------HDGP-VFSMHALE-K-G--FVT-G--GKD------G--IVALWD WD17 877-914 AP------G----SKG--LL----------LEDN-PSIRAISL-G-HGHILV-G--TKN------G--EILEVD WD18 918-956 -P------I----TLL--VQG-H-------MEGE-VWGLATHP-H-LPLCAT-V--SDD------K--TLRIWD WD19 960-998 -S------H----CML--AVR-K-------LKKG-GRCCCFSP-D-GKALAV-G--LND------G--SFLIVN WD20 1002-1041 -L------E----DLV--SFH-H-------RKDI-ISEIRFSPVT-GKYLAV-A--SGD------S--FVDIYN WD21 1045-1084 -S------K----RVG--VCK-G-------SMNY-ITHLDWDK-RVGKLLQV-N--TSA------K--EQLFFE WD22 1114-1152 -C------E----GIW--PVV-S-------EVTE-VTAACLSY-D-KKLLAT-G--DDL------G--YVKLFK WD23 1159-1198 -Y------A----KFK--RYM-A-------HSTH-VTNVRWSH-D-DSLLASVG--GSD------T--CLMIWN WD24 1201-1239 -T------E----SCR--EAR-Q-------CDSE-ESDIESED-D-GGYDSD-V--TRE------N--EIVYVI WD25 1249-1294 MTGVKPHLQ----QKE--PTV-D-------ESDG-FLEMCRPP-V-SRAPPM-P--EKL------Q--TNNVGK WD26 1295-1341 KK------R----PIE--DLV-LELVFGYRGNDC-RNNVHYLN-E-GADIIY-H--TAS------I--GVVLNL WD27 1352-1393 HS------D--DILCL--TIN-Q-------HPKF-PNVVATGQ-V-GDTGDM-S--ATS------P--SIHVWE WD28 1397-1438 -K------Q----TLS--VLRCH-------HSRG-VCSVSFSA-T-GKLLLS-V--GLD----PQH--TLTIWK WD29 1442-1480 -G------A----KVA--SRA-G-------HTQR-IFVAEFRP-D-SDTQFV-S--VGI------K--HVRFWT WD30 1491-1528 -G------V----LSS--IED-A-------RMQT-MLSVAFGA-N-NLTF-T-G--TIS------G--DVCVWK WD31 1529-1575 EH------I----LVR-IVAK-A-------HTGP-VFTMYTTL-R-DGLIVT-G--GKERPSKEGG--ALKLWD WD32 1576-1619 QE--LKRCR----AFR--LET-G-------QVID-CVRSVCRG-K-GKILVG-T--RNA------E--IIEVGE WD33 1622-1661 -A------A----CNI--LVN-G-------HMDGPVWGLGAHP-T-RDVFLS-A--AED------G--TVRLWD WD34 1662-1702 IS------Q---RKML--NKV-N-------LGHP-ARTVSYSP-E-GDMVAI-G--MKN------G--EFIILL WD35 1706-1744 -L------K----IWG--KKR-D-------RRSA-IQDIRFSP-D-SRYLAV-G--SSE------N--AVDFYD WD36 1754-1789 --------------IN--CCR-D-------IPSF-VMQMDFSA-D-SCFVQL-S--TGA------Y--KRVVYE WD37 1815-1856 GD------E----VVGIWSRN-T-------DKAD-VTCACVSH-S-GINIVT-G--DDF------G--MVKLFD WD38 1863-1902 -F------A----KHK--RFL-G-------HSAH-LTNVRFTN-G-DRFVVSAG--GDD------R--SLFVWR WD40 Template TG------E----CLR--TLS-G-------HTSG-VTSVSFSP-D-GKRLVS-G--SED------G--TIKVWD |
|
Copyright © 2012-2015 Ziding Zhang's Lab, China Agricultural University Powered and maintained by Chuan Wang Last modified: 09 Jan 2019 |