WDRR: WD40 Repeat Recognition
| [HOME] [JOB LIST] [PRE-CALCULATED RESULTS] [VISITORS] [ABOUT] |
| Pre-calculated Results - Accession: ENSDARP00000079585 [Danio rerio] [Download result as txt] [Query sequence in FASTA]
Input parameters:
Query = ENSDARP00000079585 (Len=1866) Tmplt = WD40-str (Len=40) Method = bdp Gap-open = 50 Gap-extend = 3 Database = /home/pub/blastdb/nr90 E-value = 0.001 Iteration = 3Query sequence (ENSDARP00000079585, Len=1866): (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) 10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
Query 1 MADKTAPRCQLRLEWIHGYRGHQCRNNLYYTAGKEIVYFVAGVGVVYNTREHTQKFYLGHNDDIISLALHPDKIQVATGQ 80
WD40-rpts (___________________WD 1________
Query 81 VGKDPYICVWDSYALTTVSILRDVHTHGVACLAFDADGQRLASVGLDAKNTVCVWDWKKGRVLATATGHSDRIFDISWDP 160
WD40-rpts __________) (__________________WD 2_________________) (_________________WD
Query 161 FLPHRLVSCGVKHIKFWALCGNALTPKRGIFGKTGDLQTILCLATAKEEVTYSGALNGDIYVWKGLNLTRTIQAAHGAGI 240
WD40-rpts 3________________) (__________________WD 4_________________)(_______________
Query 241 FSMHSCEEGFATGGRDGCVRLWDVDFKPITKIDLREAEQGYKGLSIRSVCWRADRILAGTQDSEIFEVMVRDRDKPLLIM 320
WD40-rpts __WD 5________________)(_____________________WD 6____________________) (______
Query 321 QGHSEGELWALDLHPKQPLAVTGSDDRSVRLWSLSEHTLIARCNMEEAVRSVSFINDGSQLALGMKDGSFTVLRVRYFSL 400
WD40-rpts ___________WD 7_________________)(__________________WD 8_________________) (__
Query 401 RSRPICLNQAYFTHLNKYNSGTIGQKHFHEEITINENLKLNRTNQNYIYIYIYMFMYVCVYCNLMLKSMHPCSPKDIMWI 480
WD40-rpts _______________WD 9________________)
Query 481 KRFSCSILLWMFVVKPGNVLLCVVSYCSEAYADSNSGESDSDLSDVSELDSDIEQEAQISYERQVYKEDLPQLKKKLAGS 560
WD40-rpts
Query 561 LKRQKAPDEGLRLQFVHGYRGCDCRNNLFYTQAGEVVYHVAAVAVVYNRQQHTQRFYLGHDDDILSLTVHPLKDYVASGQ 640
WD40-rpts (__________________WD10_______
Query 641 VGRDPAIHVWDIQTLKCLSLLKGQHQRGVCALEFTADGKSLVSVGIDEEHCIVIWDWKKGEKLAKSRGHKDKIFVVKGNP 720
WD40-rpts __________) (__________________WD11__________________) (_________________WD1
Query 721 FRMDKLVTVGMKHIKFWLHTVVNAGGGLTFRRGIFGNLGKQETMMSACYGRSEDLVFSGATTGDVYIWKDTTLMKSIKAH 800
WD40-rpts 2________________) (______________WD13______________)(__________
Query 801 DGPVFAMCSLDKIYVTGGKDGVVELWDDMFERCLKTYAIKRAALSPSSKGLLLEDNPSIRAITLGHGHILVGTKNGEILE 880
WD40-rpts ______WD14________________) (________________WD15_______________
Query 881 IDKSGPMTLLVQGHMEGEVWGLAAHPLLPICATVSDDKTLRIWELSANHRMVAVRKLKKVGGRCCAFSPDGKALAVGLND 960
WD40-rpts _) (_________________WD16________________)(___________________WD17____________
Query 961 GSFLVVNADTLEDMVTFHHRKEIISDIRFSQDAGKYLAVASHDSFVDIYNVLTSKRVGICKGASSCITHVDWDVRGKLLQ 1040
WD40-rpts ______) (_________________WD18_________________) (_________________WD19_____
Query 1041 VNTGAKEQLFFEAPRGRRQTISSLEFEKMEWATWTSVLGGTCEGIWPTLSLVNASSLTKDKKLLATGDDFGFVKLFSFPS 1120
WD40-rpts ___________) (_________________WD20________________)
Query 1121 KGQFAKFKKYVAHSANVTNVRWSNDDAMLLSVGGADTALMIWAREGIGPRESKVVDSEESDDDAEEDGGYDSDVAREKNM 1200
WD40-rpts (_________________WD21_________________) (_________________WD22_________
Query 1201 DYTTKIYAVSIRQMTGVKPHQQQKEILVDERPPVSRAAPLPDKLVKNNITKKKKTVEELSLDHVFGYRGFDCRNNLHYLN 1280
WD40-rpts _______) (_________________WD23_________________) (___________
Query 1281 DGADIIFHTAAAAVIHSLSAGTQSFYLEHTDDILCLTVNQHPKYQNIIATGQIGQCVTFTGLAPSIHVWDAMSKQTMSVL 1360
WD40-rpts ________WD24__________________)(_________________WD25________________)(_________
Query 1361 RCAHAKGIGYVNFSATGKLLLSVGVDPEHTITVWRWQEGSRVCSKGGHTDRIFVVEFRPDSDTQFVSVGIKHIKFWTLVG 1440
WD40-rpts ___________WD26___________________) (_________________WD27________________)
Query 1441 GSLLYKKGVIGAVEDGHMQTMLSVAFGANNLTFTGAINGDVFVWREHFLVRVVAKAHTGPVFTMYTTLRDGLIVTGGKER 1520
WD40-rpts (________________WD28________________)(_____________________WD29_________
Query 1521 PTKEGGAVKLWDQEMKRCRAFQLETGLPVEIVRSVCRGKGKILVGTKDGEIMEVGEKNAASNTIINGHTQGRIWGLATHP 1600
WD40-rpts ___________)(___________________WD30___________________) (_________________WD31
Query 1601 SKDVFISASDDGTIRFWDLADKKLLNKVSLGHPAKCTAYSPNGEMVSIGMENGEFIVLLVNSLTVWGKKRDRSVAIQDIR 1680
WD40-rpts _________________)(__________________WD32_________________) (_________________
Query 1681 FSSDNRLLAVGSVESAVDFYDLTLGPSLNRIGYCKDIPGFVIQLDFSADSKYIEQVSTGSYKRQIHEVPSGKLVSDQALM 1760
WD40-rpts WD33________________) (________________WD34_______________)
Query 1761 DRITWASWTSVLGDEVLGIWPRNAEKADVNCACVSHAGINVVTGDDFGLVKLFDFPCSEKFAKHKRYFGHSAHVTNIRFS 1840
WD40-rpts (_____________________WD35____________________) (_________________WD
Query 1841 YDDKYVISVGGNDCSVFVWRCVSSPF 1866
WD40-rpts 36_________________)
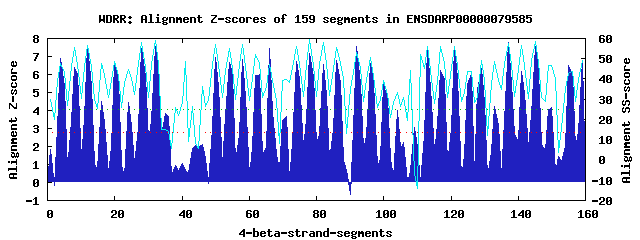 36 WD40-repeats found in QUERY=ENSDARP00000079585 (Len=1866): (Red - Z-score>4.0) Rpts Segment Boundary Z-score Score P-value Identities SS-score WD 1: Seg 4 49-91 6.91 400.11 0.00025 32.5% 47.66 WD 2: Seg 8 96-136 6.40 379.10 0.00048 37.5% 55.77 WD 3: Seg 12 140-178 7.60 428.46 0.00010 25.0% 56.44 WD 4: Seg 16 184-224 4.50 300.57 0.00553 25.0% 47.76 WD 5: Seg 20 225-263 6.65 389.52 0.00035 27.5% 48.54 WD 6: Seg 24 264-310 4.44 298.08 0.00597 15.0% 44.87 WD 7: Seg 28 314-353 7.46 422.91 0.00012 25.0% 57.89 WD 8: Seg 32 354-394 7.58 427.80 0.00011 35.0% 59.05 WD 9: Seg 35 398-436 3.86 274.12 0.01252 2.5% 15.12 WD10: Seg 50 611-651 6.92 400.58 0.00025 30.0% 57.11 WD11: Seg 54 655-696 6.68 390.45 0.00034 45.0% 54.99 WD12: Seg 58 700-738 6.82 396.40 0.00028 25.0% 57.00 WD13: Seg 63 756-789 6.01 363.07 0.00080 15.0% 47.83 WD14: Seg 66 790-827 7.47 423.07 0.00012 22.5% 47.25 WD15: Seg 71 845-882 3.69 266.90 0.01565 12.5% 39.91 WD16: Seg 74 886-924 6.73 392.77 0.00032 17.5% 56.13 WD17: Seg 78 925-967 7.15 409.97 0.00019 30.0% 59.44 WD18: Seg 82 971-1010 6.58 386.46 0.00039 25.0% 58.23 WD19: Seg 86 1014-1052 6.78 394.92 0.00030 17.5% 55.60 WD20: Seg 92 1079-1117 7.56 427.14 0.00011 25.0% 53.18 WD21: Seg 96 1124-1163 6.63 388.60 0.00036 27.5% 50.45 WD22: Seg100 1170-1208 5.43 338.97 0.00168 15.0% 39.40 WD23: Seg104 1210-1249 4.01 280.35 0.01033 7.5% 33.14 WD24: Seg109 1269-1311 3.08 242.02 0.03356 15.0% -6.46 WD25: Seg113 1312-1350 7.44 422.16 0.00013 17.5% 55.81 WD26: Seg117 1351-1395 6.21 371.33 0.00062 35.0% 56.24 WD27: Seg121 1399-1437 7.50 424.58 0.00012 27.5% 56.11 WD28: Seg126 1448-1485 6.06 364.81 0.00075 25.0% 43.87 WD29: Seg129 1486-1532 6.31 375.30 0.00054 25.0% 49.30 WD30: Seg133 1533-1576 4.23 289.25 0.00785 15.0% 48.72 WD31: Seg137 1579-1618 7.77 435.71 8.3e-05 32.5% 57.69 WD32: Seg141 1619-1659 6.17 369.65 0.00065 20.0% 56.37 WD33: Seg145 1663-1701 7.56 427.13 0.00011 17.5% 58.72 WD34: Seg150 1711-1747 4.19 287.76 0.00822 20.0% 46.66 WD35: Seg155 1768-1814 6.48 382.24 0.00044 22.5% 42.65 WD36: Seg159 1821-1860 6.76 394.08 0.00030 32.5% 48.94Multiple sequence alignment of 36 WD40-repeats: (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) WD 1 49-91 TR----E--HT---QK-------F-YLG--------HNDD-IISLALHPD-KI--QVAT-GQVG--KD------P--YICVWD WD 2 96-136 -T----T---V---SI-------L-RDV--------HTHG-VACLAFDAD-GQ--RLAS-V--G--LD------AKNTVCVWD WD 3 140-178 -G----R---V---LA-------T-ATG--------HSDR-IFDISWDPF-LP--HRLV-S--C--GV------K--HIKFWA WD 4 184-224 -L----T---P---KR-------G-IFG--------KTGD-LQTILCLAT-AKEEVTYS-G--A--LN------G--DIYVWK WD 5 225-263 GL----N---L---TR-------TIQAA--------HGAG-IFSMHSCEE-G----FAT-G--G--RD------G--CVRLWD WD 6 264-310 VD----F---K---PI-------T-KIDLREAEQGYKGLS-IRSVCWRAD--R--ILAG-T--Q--DS------E--IFEVMV WD 7 314-353 -D----K---P---LL-------I-MQG--------HSEGELWALDLHPK-QP--LAVT-G--S--DD------R--SVRLWS WD 8 354-394 LS----E--HT---LI-------A-RCN--------MEEA-VRSVSFIND-GS--QLAL-G--M--KD------G--SFTVLR WD 9 398-436 -F----S---L---RS-------R-PIC--------LNQA-YFTHLNKYN-SG--TIGQ-K--H--FH------E--EITINE WD10 611-651 QH--------T---QR-------F-YLG--------HDDD-ILSLTVHPL-KD--YVAS-G--QVGRD------P--AIHVWD WD11 655-696 -L----K---C---LS-------L-LKG-------QHQRG-VCALEFTAD-GK--SLVS-V--G--ID----EEH--CIVIWD WD12 700-738 -G----E---K---LA-------K-SRG--------HKDK-IFVVKGNPF-RM--DKLV-T--V--GM------K--HIKFWL WD13 756-789 -----------------------G-NLG--------KQET-MMSACYGRS-ED--LVFS-G--A--TT------G--DVYIWK WD14 790-827 DT----T---L---MK-------S-IKA--------HDGP-VFA--MCSL-DK--IYVT-G--G--KD------G--VVELWD WD15 845-882 SP----S---S---KG-------L-LLE--------DNPS-IRAI--TLG-HG--HILV-G--T--KN------G--EILEID WD16 886-924 -P----M---T---LL-------V-QGH--------MEGE-VWGLAAHPL-LP--ICAT-V--S--DD------K--TLRIWE WD17 925-967 LS-ANHR---M---VA-------V-RKL--------KKVG-GRCCAFSPD-GK--ALAV-G--L--ND------G--SFLVVN WD18 971-1010 -L----E---D---MV-------T-FHH--------RKEI-ISDIRFSQDAGK--YLAV-A--S--HD------S--FVDIYN WD19 1014-1052 -S----K---R---VG-------I-CKG--------ASSC-ITHVDWDVR-GK--LLQV-N--T--GA------K--EQLFFE WD20 1079-1117 -G----G---T---CE-------G-IWP--------TLSL-VNASSLTKD-KK--LLAT-G--D--DF------G--FVKLFS WD21 1124-1163 -F----A---K---FK-------K-YVA--------HSAN-VTNVRWSND-DA--MLLSVG--G--AD------T--ALMIWA WD22 1170-1208 -R----E---S---KV-------V-DSE--------ESDD-DAEEDGGYD-SD--VARE-K--N--MD------Y--TTKIYA WD23 1210-1249 SI----R---Q---MT-------G-VKP--------HQQQ-KEILVDERP-PV--SRAA-P--L--PD------K--LVKNNI WD24 1269-1311 GF----D---CRNNLH-------Y-LND--------GADI-IFHTAAAAV-IH--SLSA-G--T--QS------F--YLEHTD WD25 1312-1350 -D----I---L---CL-------T-VNQ--------HPKY-QNIIATGQI-GQ--CVTF-T--G--LA------P--SIHVWD WD26 1351-1395 AM----SKQTM---SV-------L-RCA--------HAKG-IGYVNFSAT-GK--LLLS-V--G--VD----PEH--TITVWR WD27 1399-1437 -G----S---R---VC-------S-KGG--------HTDR-IFVVEFRPD-SD--TQFV-S--V--GI------K--HIKFWT WD28 1448-1485 -G----V---I---GA-------V-EDG--------HMQT-MLSVAFGAN-N---LTFT-G--A--IN------G--DVFVWR WD29 1486-1532 EH----F---L---VR------VV-AKA--------HTGP-VFTMYTTLR-DG--LIVT-G--G--KERPTKEGG--AVKLWD WD30 1533-1576 QEMKRCR---A---FQ-------L-ETG--------LPVE-IVRSVCRGK-GK--ILVG-T--K--DG------E--IMEVGE WD31 1579-1618 -A----A---S---NT-------I-ING--------HTQGRIWGLATHPS-KD--VFIS-A--S--DD------G--TIRFWD WD32 1619-1659 LA----D--KK---LL-------N-KVS--------LGHP-AKCTAYSPN-GE--MVSI-G--M--EN------G--EFIVLL WD33 1663-1701 -L----T---V---WG-------K-KRD--------RSVA-IQDIRFSSD-NR--LLAV-G--S--VE------S--AVDFYD WD34 1711-1747 IG----Y---C---------------KD--------IPGF-VIQLDFSAD-SK--YIEQ-V--S-TGS------Y--KRQIHE WD35 1768-1814 WT----S---V---LGDEVLGIWP-RNA--------EKAD-VNCACVSHA-GI--NVVT-G--D--DF------G--LVKLFD WD36 1821-1860 -F----A---K---HK-------R-YFG--------HSAH-VTNIRFSYD-DK--YVISVG--G--ND------C--SVFVWR WD40 Template TG----E---C---LR-------T-LSG--------HTSG-VTSVSFSPD-GK--RLVS-G--S--ED------G--TIKVWD |
|
Copyright © 2012-2015 Ziding Zhang's Lab, China Agricultural University Powered and maintained by Chuan Wang Last modified: 09 Jan 2019 |