WDRR: WD40 Repeat Recognition
| [HOME] [JOB LIST] [PRE-CALCULATED RESULTS] [VISITORS] [ABOUT] |
| Pre-calculated Results - Accession: ENSDARP00000068125 [Danio rerio] [Download result as txt] [Query sequence in FASTA]
Input parameters:
Query = ENSDARP00000068125 (Len=911) Tmplt = WD40-str (Len=40) Method = bdp Gap-open = 50 Gap-extend = 3 Database = /home/pub/blastdb/nr90 E-value = 0.001 Iteration = 3Query sequence (ENSDARP00000068125, Len=911): (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) 10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
Query 1 MAFITKKDRSKERFSLLLLDLEEQYFEQHIAYNLTNRGPETNRRTNGSLKICSKSVIFEPDDAVKPILKIPLKDCSEIAA 80
WD40-rpts
Query 81 VEETAHNPFIESKPACIVIKTKQIYQIKEENVVAPYKYDRGEKKITFQLGSPIKTEDVVQILLQLHRASRLDKQSDQTAM 160
WD40-rpts
Query 161 IAAILQSRLARTRFDKNSFQNVTELPHMECEAEMVSPLVTNAGHVCITDCNLYFQPLNSYPDSVVQIGLHSVRRIYKRRH 240
WD40-rpts
Query 241 GLRPLGLEVFCTENDLCSDIYLKFYKTKDRDELYYYIATFLENHVAEHTAESYMLQWQRGHISNYQYLLHLNNLADRSVN 320
WD40-rpts
Query 321 DLSQYPVFPWVISDYSSTQLDLLNPASFRDLSKPIGALNKERLERLLERYRDMPEPRFMYGSHYSSPGYVLFYLVRVAPE 400
WD40-rpts
Query 401 HMLCLQNGRFDHADRMFNSIGETWKNCLEGGTDFKELIPEFYGSDSSFLRNLLGLDLGRRQGGGRVENVELPPWASDPDD 480
WD40-rpts
Query 481 FLQKMCLALESQYVSEHLHEWIDLIFGYKQRGSEAVASHNVFHPLTYEGGVDCDSIEDPDQKIAMLTQILEFGQTPRQLF 560
WD40-rpts
Query 561 VTPHPQRITPRFYNLSTTPSLSSSELSPVSPCMESFEDLTEESRKMAWSNMNKLVLQSCHKIHKEAVTGVAVTLSGDSIF 640
WD40-rpts (___________________WD 1_______
Query 641 TTSQDSTLKMFSKENGLQRSVSFSNMALSSCLMLPDDKIVVCSSWDNNVYFYSIAYGRRQDTLMGHDDAVSDICWRDDKL 720
WD40-rpts ___________)(__________________WD 2_________________) (________________WD 3___
Query 721 YTASWDSTVKVWECVAADISSNKRTQFDPLAEFEHEAGVNTLDLSPAGTLLATGTKEGTLTVWDLASPLPVNQLTCHSGK 800
WD40-rpts ____________)(_______________________WD 4______________________) (____________
Query 801 IHQVAFSPDSRHILSVGEDSCLMVTDVQTGMLIATIPAEQEQRCFCWDGNTVLSGGISGDLSVWNLLNSKVVHKIPAAHS 880
WD40-rpts _____WD 5________________)(_________________WD 6________________)(______________
Query 881 GAVTCMWMNEQCSSIITGGLDKQIILWKPQY 911
WD40-rpts _____WD 7__________________)
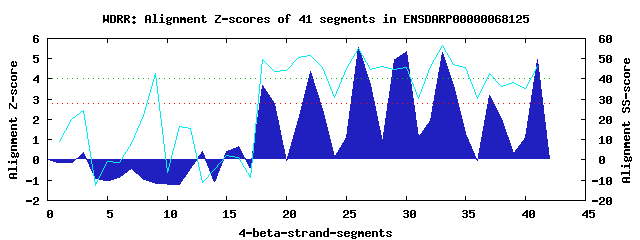 7 WD40-repeats found in QUERY=ENSDARP00000068125 (Len=911): (Red - Z-score>4.0) Rpts Segment Boundary Z-score Score P-value Identities SS-score WD 1: Seg 18 610-652 3.69 266.97 0.01561 25.0% 49.80 WD 2: Seg 22 653-693 4.37 295.06 0.00656 25.0% 51.70 WD 3: Seg 26 697-733 5.57 344.95 0.00140 35.0% 55.27 WD 4: Seg 30 734-784 5.37 336.47 0.00182 32.5% 45.91 WD 5: Seg 33 788-826 5.31 333.88 0.00197 30.0% 56.71 WD 6: Seg 37 827-865 3.18 246.16 0.02958 20.0% 42.95 WD 7: Seg 41 866-908 4.95 319.18 0.00311 17.5% 46.06Multiple sequence alignment of 7 WD40-repeats: (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) WD 1 610-652 NMNK-----------LVLQSCHKIHKEAVTGVAVTLSGDSIFTTSQDSTLKMFS WD 2 653-693 KENG-----------L--QRSVSFSNMALSSCLMLPDDKIVVCSSWDNNVYFYS WD 3 697-733 -GRR-----------Q---DTLMGHDDAVSDICWRDD--KLYTASWDSTVKVWE WD 4 734-784 CVAADISSNKRTQFDP---LAEFEHEAGVNTLDLSPAGTLLATGTKEGTLTVWD WD 5 788-826 -PLP-----------V---NQLTCHSGKIHQVAFSPDSRHILSVGEDSCLMVTD WD 6 827-865 VQTG-----------M----LIATIPAEQEQRCFCWDGNTVLSGGISGDLSVWN WD 7 866-908 LLNS-----------KVVHKIPAAHSGAVTCMWMNEQCSSIITGGLDKQIILWK WD40 Template TGEC-----------L---RTLSGHTSGVTSVSFSPDGKRLVSGSEDGTIKVWD |
|
Copyright © 2012-2015 Ziding Zhang's Lab, China Agricultural University Powered and maintained by Chuan Wang Last modified: 09 Jan 2019 |