WDRR: WD40 Repeat Recognition
| [HOME] [JOB LIST] [PRE-CALCULATED RESULTS] [VISITORS] [ABOUT] |
| Pre-calculated Results - Accession: CC4.3 [Caenorhabditis elegans] [Download result as txt] [Query sequence in FASTA]
Input parameters:
Query = CC4.3 (Len=510) Tmplt = WD40-str (Len=40) Method = bdp Gap-open = 50 Gap-extend = 3 Database = /home/pub/blastdb/nr90 E-value = 0.001 Iteration = 3Query sequence (CC4.3, Len=510): (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) 10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
Query 1 MSSIEIESSDVIRLIEQFLKESNLHRTLAILQEETNVSLNTVDSIDGFCNEITSGNWDNVLKTVQSLKLPAKKLIDLYEH 80
WD40-rpts (_____________WD 1____________) (___________
Query 81 VIIELVELRELATARLVARQTDPMILLKQIDPDRFARLESLINRPYFDGQEVYGDVSKEKRRSVIAQTLSSEVHVVAPSR 160
WD40-rpts _______WD 2_________________) (___________
Query 161 LLSLLGQSLKWQLHQGLLPPGTAIDLFRGKAAQKEQIEERYPTMMARSIKFSTKSYPESAVFSPDANYLVSGSKDGFIEV 240
WD40-rpts ____________WD 3______________________) (_________________WD 4_______________
Query 241 WNYMNGKLRKDLKYQAQDNLMMMDAAVRCISFSRDSEMLATGSIDGKIKVWKVETGDCLRRFDRAHTKGVCAVRFSKDNS 320
WD40-rpts _) (_________________WD 5________________) (_________________WD 6___
Query 321 HILSGGNDHVVRVHGMKSGKCLKEMRGHSSYITDVRYSDEGNHIISCSTDGSIRVWHGKSGECLSTFRVGSEDYPILNVI 400
WD40-rpts ______________) (_________________WD 7________________) (___________________
Query 401 PIPKSDPPQMIVCNRSNTLYVVNISGQVVRTMTSGKREKGDFINCILSPKGEWAYAIAEDGVMYCFMVLSGTLETTLPVT 480
WD40-rpts WD 8__________________)(___________________WD 9___________________) (_________
Query 481 ERLPIGLAHHPHQNLIASYAEDGLLKLWTD 510
WD40-rpts ________WD10________________)
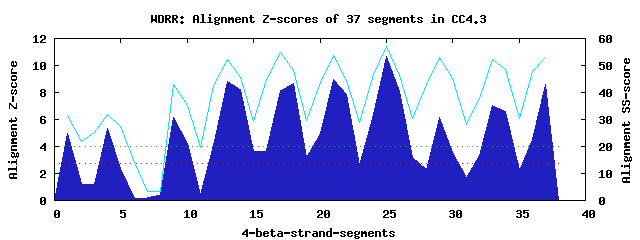 10 WD40-repeats found in QUERY=CC4.3 (Len=510): (Red - Z-score>4.0) Rpts Segment Boundary Z-score Score P-value Identities SS-score WD 1: Seg 1 35-65 4.97 320.11 0.00302 17.5% 31.53 WD 2: Seg 4 69-109 5.32 334.43 0.00194 12.5% 31.73 WD 3: Seg 9 149-199 6.13 367.96 0.00068 10.0% 43.09 WD 4: Seg 13 204-242 8.83 479.35 2.2e-05 40.0% 52.39 WD 5: Seg 18 254-292 8.70 474.24 2.5e-05 37.5% 48.11 WD 6: Seg 21 296-335 8.94 484.09 1.9e-05 40.0% 53.77 WD 7: Seg 25 339-377 10.70 556.47 2.0e-06 42.5% 56.89 WD 8: Seg 29 381-423 6.16 368.95 0.00066 22.5% 53.05 WD 9: Seg 33 424-467 7.05 405.79 0.00021 15.0% 52.04 WD10: Seg 37 471-509 8.62 470.71 2.8e-05 25.0% 53.15Multiple sequence alignment of 10 WD40-repeats: (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) WD 1 35-65 TN----VSLNTV-DS---------IDGFCNEI-----------------TSGNWDNVLKTVQ WD 2 69-109 -L----PAKKLI-DL---------YEHVIIELVELR----ELATAR--LVARQTDPMILLKQ WD 3 149-199 LS----SEVHVV-APSRLLSLLGQSLKWQLHQGLLP----PGT--AIDLFRGKAAQKEQIEE WD 4 204-242 -M----MARSIK-FS---------TKSYPESAVFSP----DAN--Y--LVSGSKDGFIEVWN WD 5 254-292 -Y----QAQDNL-MM---------MDAAVRCISFSR----DSE--M--LATGSIDGKIKVWK WD 6 296-335 -G----DCLRRFDRA---------HTKGVCAVRFSK----DNS--H--ILSGGNDHVVRVHG WD 7 339-377 -G----KCLKEM-RG---------HSSYITDVRYSD----EGN--H--IISCSTDGSIRVWH WD 8 381-423 -G----ECLSTF-RV---------GSEDYPILNVIPIPKSDPP--Q--MIVCNRSNTLYVVN WD 9 424-467 ISGQVVRTMTSG-KR---------EKGDFINCILSP----KGE--W--AYAIAEDGVMYCFM WD10 471-509 -G----TLETTL-PV---------TERLPIGLAHHP----HQN--L--IASYAEDGLLKLWT WD40 Template TG----ECLRTL-SG---------HTSGVTSVSFSP----DGK--R--LVSGSEDGTIKVWD |
|
Copyright © 2012-2015 Ziding Zhang's Lab, China Agricultural University Powered and maintained by Chuan Wang Last modified: 09 Jan 2019 |