WDRR: WD40 Repeat Recognition
| [HOME] [JOB LIST] [PRE-CALCULATED RESULTS] [VISITORS] [ABOUT] |
| Pre-calculated Results - Accession: AT4G34380.1 [Arabidopsis thaliana] [Download result as txt] [Query sequence in FASTA]
Input parameters:
Query = AT4G34380.1 (Len=495) Tmplt = WD40-str (Len=40) Method = bdp Gap-open = 50 Gap-extend = 3 Database = /home/pub/blastdb/nr90 E-value = 0.001 Iteration = 3Query sequence (AT4G34380.1, Len=495): (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) 10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
Query 1 MRNETGGTMVVDLQTDPYNNIHSRHNLSYDLSEGPKPRPKFGNISKTDHDEIFPPDDPIIINSNVTHQRFSSVSASTMSS 80
WD40-rpts (________
Query 81 GPASGEGSPCVMSPWARVSPPWGADFNEDNVLETNGLIGSIVRKEGHIYSLAASGDLLYTGSDSKNIRVWKNLKEHAGFK 160
WD40-rpts _____WD 1_____________) (________________WD 2_______________)(________
Query 161 SSSGLIKAIVIFGDRIFTGHQDGKIRIWKVSKRKPGKHKRVGTLPTFKSMVKSSVNPKHFMEVRRNRNSVKTKHNDAVSS 240
WD40-rpts ________WD 3________________)(______________WD 4______________)(________________
Query 241 LSLDVELGLLYSSSWDTTIKVWRIADSKCLESIHAHDDAINSVMSGFDDLVFTGSADGTVKVWKRELQGKGTKHTLAQVL 320
WD40-rpts _WD 5_________________) (________________WD 6________________) (______
Query 321 LKQENAVTALAVKSQSSIVYCGSSDGLVNYWERSKRSFTGGILKGHKSAVLCLGIAGNLLLSGSADKNICVWRRDPSDKS 400
WD40-rpts ___________WD 7________________)(__________________WD 8_________________)
Query 401 HQCLSVLTGHMGPVKCLAVEEERACHQGAKASVAEGDRKWIIYSGSLDKSVKVWRVSERMATWKEMDEPAASSERKSSSS 480
WD40-rpts (_______________WD 9_______________)
Query 481 PHEGSGAWSSRNVEK 495
WD40-rpts
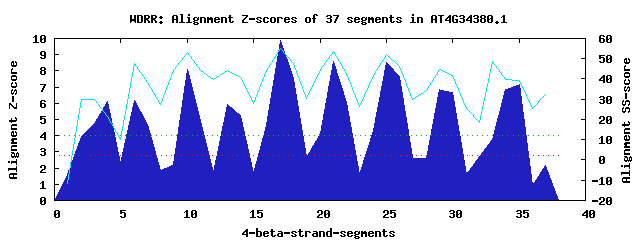 9 WD40-repeats found in QUERY=AT4G34380.1 (Len=495): (Red - Z-score>4.0) Rpts Segment Boundary Z-score Score P-value Identities SS-score WD 1: Seg 4 72-103 6.14 368.16 0.00068 17.5% 21.45 WD 2: Seg 6 115-151 6.18 369.94 0.00064 20.0% 47.50 WD 3: Seg 10 152-189 8.11 449.57 5.4e-05 17.5% 52.93 WD 4: Seg 14 190-223 5.26 332.03 0.00209 12.5% 40.56 WD 5: Seg 17 224-263 9.88 522.97 5.6e-06 32.5% 55.14 WD 6: Seg 21 267-304 8.55 467.67 3.1e-05 35.0% 53.65 WD 7: Seg 25 314-352 8.50 465.64 3.3e-05 20.0% 52.03 WD 8: Seg 29 353-393 6.84 397.40 0.00027 35.0% 44.55 WD 9: Seg 35 420-455 7.14 409.54 0.00019 27.5% 39.18Multiple sequence alignment of 9 WD40-repeats: (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) WD 1 72-103 -----------SVSASTMSSGPASGEGSPCVMSPWARVSPPWG WD 2 115-151 -NG---LIGSIVRKEGHIYSL--AASGDLLYTGSDSKNIRVWK WD 3 152-189 NLK---EHAGFKSSSGLIKAIVIFGD--RIFTGHQDGKIRIWK WD 4 190-223 ---------VSKRKPGKHKRVGTLPTFKSMVKSSVNPKHFMEV WD 5 224-263 RRN---RNSVKTKHNDAVSSLSLDVELGLLYSSSWDTTIKVWR WD 6 267-304 -SK---CLESIHAHDDAINSVMSGFDD-LVFTGSADGTVKVWK WD 7 314-352 -HT---LAQVLLKQENAVTALAVKSQSSIVYCGSSDGLVNYWE WD 8 353-393 RSKRSFTGGILKGHKSAVLCLGIA--GNLLLSGSADKNICVWR WD 9 420-455 EEE-------RACHQGAKASVAEGDRKWIIYSGSLDKSVKVWR WD40 Template TGE---CLRTLSGHTSGVTSVSFSPDGKRLVSGSEDGTIKVWD |
|
Copyright © 2012-2015 Ziding Zhang's Lab, China Agricultural University Powered and maintained by Chuan Wang Last modified: 09 Jan 2019 |