WDRR: WD40 Repeat Recognition
| [HOME] [JOB LIST] [PRE-CALCULATED RESULTS] [VISITORS] [ABOUT] |
| Pre-calculated Results - Accession: AT4G05410.1 [Arabidopsis thaliana] [Download result as txt] [Query sequence in FASTA]
Input parameters:
Query = AT4G05410.1 (Len=504) Tmplt = WD40-str (Len=40) Method = bdp Gap-open = 50 Gap-extend = 3 Database = /home/pub/blastdb/nr90 E-value = 0.001 Iteration = 3Query sequence (AT4G05410.1, Len=504): (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) 10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
Query 1 MKYNNEKKKGGSFKRGGKKGSNERDPFFEEEPKKRRKVSYDDDDIESVDSDAEENGFTGGDEDGRRVDGEVEDEDEFADE 80
WD40-rpts
Query 81 TAGEKRKRLAEEMLNRRREAMRREREEADNDDDDDEDDDETIKKSLMQKQQEDSGRIRRLIASRVQEPLSTDGFSVIVKH 160
WD40-rpts (_________
Query 161 RRSVVSVALSDDDSRGFSASKDGTIMHWDVSSGKTDKYIWPSDEILKSHGMKLREPRNKNHSRESLALAVSSDGRYLATG 240
WD40-rpts ________WD 1________________) (_________________WD 2_______
Query 241 GVDRHVHIWDVRTREHVQAFPGHRNTVSCLCFRYGTSELYSGSFDRTVKVWNVEDKAFITENHGHQGEILAIDALRKERA 320
WD40-rpts _________) (_________________WD 3________________) (________________WD 4____
Query 321 LTVGRDRTMLYHKVPESTRMIYRAPASSLESCCFISDNEYLSGSDNGTVALWGMLKKKPVFVFKNAHQDIPDGITTNGIL 400
WD40-rpts ____________) (________________WD 5________________) (________________
Query 401 ENGDHEPVNNNCSANSWVNAVATSRGSDLAASGAGNGFVRLWAVETNAIRPLYELPLTGFVNSLAFAKSGKFLIAGVGQE 480
WD40-rpts ___________WD 6___________________________) (_________________WD 7__________
Query 481 TRFGRWGCLKSAQNGVAIHPLRLA 504
WD40-rpts ______)
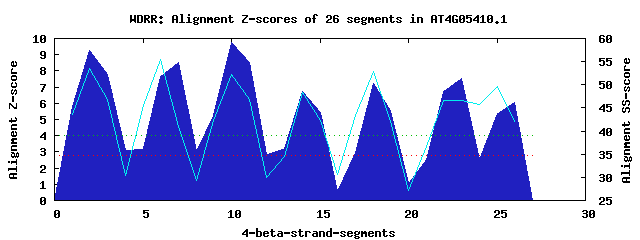 7 WD40-repeats found in QUERY=AT4G05410.1 (Len=504): (Red - Z-score>4.0) Rpts Segment Boundary Z-score Score P-value Identities SS-score WD 1: Seg 2 151-189 9.27 497.58 1.2e-05 37.5% 53.61 WD 2: Seg 7 212-250 8.54 467.60 3.1e-05 22.5% 40.95 WD 3: Seg 10 254-292 9.74 516.80 6.7e-06 35.0% 52.30 WD 4: Seg 14 296-333 6.75 393.31 0.00031 15.0% 48.24 WD 5: Seg 18 336-373 7.21 412.31 0.00017 27.5% 52.82 WD 6: Seg 23 384-443 7.50 424.53 0.00012 22.5% 46.60 WD 7: Seg 26 449-487 6.08 365.85 0.00073 25.0% 42.13Multiple sequence alignment of 7 WD40-repeats: (BLUE - beta-strands, RED - alpha-helices, predicted by PSIPRED) WD 1 151-189 -T--------------------DGFSVIVKHRRSVVSVALSDDDSRGFSASKDGT-IMHWD WD 2 212-250 -K--------------------LREPRNKNHSRESLALAVSSDGRYLATGGVDRH-VHIWD WD 3 254-292 -R--------------------EHVQAFPGHRNTVSCLCFRYGTSELYSGSFDRT-VKVWN WD 4 296-333 -K--------------------AFITENHGHQGEILAIDALRKERALTVGR-DRT-MLYHK WD 5 336-373 -E--------------------STRMIYRAPASSLESCCFISDNEYL-SGSDNGT-VALWG WD 6 384-443 KNAHQDIPDGITTNGILENGDHEPVNNNCSANSWVNAVATSRGSDLAASGAGNGF-VRLWA WD 7 449-487 IR--------------------PLYEL--PLTGFVNSLAFAKSGKFLIAGVGQETRFGRWG WD40 Template TG--------------------ECLRTLSGHTSGVTSVSFSPDGKRLVSGSEDGT-IKVWD |
|
Copyright © 2012-2015 Ziding Zhang's Lab, China Agricultural University Powered and maintained by Chuan Wang Last modified: 09 Jan 2019 |